Original Article
Haema 2022; 13(1-2):31-42
Aikaterini Bitsani, Anna Efthymiou, Maria Roumelioti, Eftychia Staikou, Aggeliki Stefanou, Danai Palaiologou, Georgios Georgiou, Georgios Boutsikas, Maria-Christina Kyrtsonis, Panayiotis Panayiotidis
Hematology section, First Department of Propaedeutic Internal Medicine, Laiko General Hospital, National and Kapodistrian University of Athens Medical School, Greece
Full PDF | ![]()
Abstract
AML development is considered a multistep process that requires the contribution of at least two classes of mutations to evolve to full-blown leukemia. Various mutations have been identified that alter the switching signal and enhance the proliferation of leukemic cells and their survival. These include mutations in the FLT3, RAS and KIT genes.
Transcription factors such as RUNX1, CEBPα and PU.1 control key genes to maintain the normal function of the hematopoietic system. Most frequent mutations are those that lead to aberrant regulation of DNA methylation and hydroxymethylation (DNMT3A, TET2 and IDH1/2).
One hundred and twenty-six (126) patients diagnosed with AML between 1995 to 2010, were retrospectively studied. (BM) specimens were stored at -80 oC . and were available for retrospective Polymerase Chain Reaction (PCR) analysis for FLT3-ITD, NPM1, and IDH1/IDH2 mutations.
When we analysed only the subgroup of patients with IDH1 mutations, we observed that it seems to have inferior outcome in terms of RFS and OS, although the statistical significance is not significant. When we analysed only the subgroup of patients with IDH2 mutations, we observed that it seems to have favorable outcome in terms of RFS and OS, although not statistically significant. When we compared IDH1 mutant patients to IDH2 ones, we observed significantly superior RFS but not statistically significant favorable outcome in OS in IDH2 mutant patients. When we compared separately IDH2 mutated patients vs wild-type, we observed that IDH2 R172 mutated patients presented the most favorable prognostic impact, followed by IDH2 R140Q mutated ones.
Key words: Acute myeloid leukemia, IDH1/IDH2 mutations
Corresponding author: Aikaterini Bitsani, Hematology Section, First Department of Propaedeutic Internal Medicine, Laiko General Hospital, National and Kapodistrian University of Athens Medical School, Greece, e-mail: kbitsani@med.uoa.gr
INTRODUCTION
Acute Myeloid Leukemia (AML) is a clonal hematological malignancy characterized by the accumulation of blast cells in the bone marrow, peripheral blood or other tissue.1 It is a heterogenous disease clinically, morphologically and genetically. The requisite blast percentage for the diagnosis of AML is at least 20% myeloblasts, monoblasts/promonocytesor megakaryoblasts in the bone marrow. A diagnosis of AML is also established when the blast percentage in the peripheral blood or bone marrow is less than 20%, if there is an associated t(8;21)(q22;q22.1), inv(16)(p13.1q22), or t(16;16)(p13.1;q22) chromosomal abnormality or a PML-RARA fusion gene. Conventional karyotype has remained one of the most important prognostic factors in AML.2
The role of genetic alterations in AML has been emphasized by the 2008 revised WHO classification of AML when, for the first time, AML with molecular genetic changes have been incorporated as provisional entities, that is, as AML with mutated NPM1 and AML with mutated CEBPA.3 In the 2016 revised WHO classification of AML a new entity is introduced, which is AML with biallelic mutations of CEBPA, and a new provisional entity constitutes AML with mutated RUNX1. According to the 2017 ELN AML Recommendations,4 as far as NPM1 and FLT3 is concerned, patients with NPM1 mutation and FLT3-ITD with a low (<0.5) allelic ratio (FLT3-ITDlow) have a similar (favorable) outcome as patients with a NPM1 mutation but no FLT3-ITD, thus, both groups are now considered favorable. In contrast, AML with wild type NPM1 and FLT3-ITD with a high (>0.5) allelic ratio (FLT3-ITDhigh) has a poor prognosis and is placed in the adverse risk group.5
AML development is considered a multistep process that requires the contribution of at least two classes of mutations to evolve to full-blown leukemia. The “two-hit model” was first described by Gilliland and Griffin.6 It includes class I mutations that activate signal transduction pathways and allow a proliferation advantage on hematopoietic cells and class II mutations that affect transcription factors and primarily block hematopoietic differentiation.7.8 Many of the newly identified genetic alterations, such as those DNMT3A, TET2, IDH1, IDH2, NPM1, ASXL1, had not been classified because the consequences of these mutations were not yet fully identified and the term “unclassified” mutations was used for this category. Recently, Metzeler et al, reported the presence of several recurrently mutated genes in acute myeloid leukemiaby performing next generation sequencing and other high-throughput techniques.9 Most frequent mutations are those that lead to aberrant regulation of DNA methylation and hydroxymethylation (DNMT3A, TET2 and IDH1/2), altered messenger RNA splicing by the U2 complex (SF3B1, SRSF2, U2AF1), modified chromatin architecture (ASXL1, EZH2 and KMT2A) and transcriptional deregulation (CEBPA, RUNX1 and WT1).10
There was a great interest concerning the role of IDH1 and IDH2 in leukemogenesis around 2010 and a number of studies tried to assess the frequencies of the above mentioned mutations and to explore their association with clinical, cytogenetic, and molecular characteristics as well as with outcome in younger adults with AML. IDH mutations are usually observed in patients with intermediate risk cytogenetics, including trisomy 8 and normal karyotype AML. Additionally, they represent early genomic events in disease pathogenesis and evolution, since they are present in dominant clones and persist even after chemotherapy or at the time of relapse or disease progression.11,12 IDH1 mutations are less common than IDH2 mutations in AML and in MDS.13,14
PATIENTS AND METHODS
One hundred and twenty-six (126) newly diagnosed AML patients (excluding M3 subtype) were retrospectively studied in one Hematology Department of Laiko General Hospital between 1995 and 2010. Medical records were reviewed for demographic and clinical data, FAB classification, karyotypic analysis, treatment strategies and outcome of the patients. Genomic DNA from pretreatment bone marrow was available for all the 126 patients and was analyzed for the FLT3, NPM1 and IDH1/IDH2 genes.
Detection of NPM1 mutations was performed by polymerase chain reaction (PCR) and fragment analysis.15,16 To amplify the exon 12 of NPM1, we used a fluorescently labeled forward primer 5-(dyeD4) TGTCTATGAAGTGTTGTGGTTCCTT-3 (Sigma-Aldrich) and a reverse primer 5-CGGTAGGGAAAGTTCTCACTCTG-3 (Sigma-Aldrich). DNA was amplified using the Hot Star Taq Plus Master Mix Kit (Qiagen). Fluorescent PCR products were subjected to capillary electrophoresis on denaturing polyacrylamide gel and analyzed by CEQ 8000 Genetic Analysis System (Beckman Coulter). Samples from healthy controls show one peak at 201bp. As all NPM1 mutations result in the insertion of 4 nucleotides, the mutated profile was defined by the presence of an additional peak at 204 bp.
Detection of FLT3-ITD mutations was performed by polymerase chain reaction (PCR) and fragment analysis as previously described.15,16 To amplify the exons 14 and 15 of FLT3, we used a forward primer 5-GCAATTTAGGTATGAAAGCCAGC-3 (Sigma-Aldrich) and fluorescently labeled reverse primer 5-(dyeD3): CTTTCAGCATTTTGACGGCAACC-3 (Sigma-Aldrich). Samples from healthy controls show one peak at 329bp. The amplicons with a size greater than that of wild type were interpreted as positive for the ITD mutation. The FLT3-ITD mutant allelic burden was calculated as the ratio of the area under the curve of mutant and wild-type alleles (mutant/total FLT3).
IDH1 mutations were detected by multiplex allele-specific PCR (AS-PCR) and agarose gel electrophoresis. To amplify the exon 4 of IDH1, we used a forward (internal control) primer 5- TGAGAAGAGGGTTGAGGAGTTCAAGT -3 (Sigma-Aldrich), a reverse primer 5-: AATGTGTTGAGATGGACGCCTATTTGT -3 (Sigma-Aldrich) and a mix of specific forward primers that are mutant specific for the following mutations R132C, R132G, R132H, R132L, R132P, R132S:
R132C: 5-TGGATGGGTAAAACCTATCATCATAGATT-3,
R132G: 5-TGGATGGGTAAAACCTATCATCATAGATG-3,
R132H: 5-GGATGGGTAAAACCTATCATCATAGGACA-3,
R132L: 5-GGATGGGTAAAACCTATCATCATAGGACT-3,
R132P: 5-GGATGGGTAAAACCTATCATCATAGGACC-3,
R132S: 5-TGGATGGGTAAAACCTATCATCATAGATA-3
DNA was amplified using the Hot StarTaq Plus Master Mix Kit (Qiagen) and PCR products were analyzed by agarose gel electrophoresis. Samples with wild type alleles give a 502-bp product. The amplicons with an extra lower band than that of wild type, were interpreted as positive for one of the specific IDH1 mutations. In order to further characterize the mutation we repeated the amplification of the DNA sample with 6 separate PCRs, using each time one of mutant specific forward primer and the common reverse.
IDH2 mutations were detected by polymerase chain reaction (PCR) and Sanger sequencing. Amplification and sequencing of IDH2 exon 4 was performed using the following primers, forward 5- GGGGTTCAAATTCTGGTTGA -3 (Sigma-Aldrich) and reverse 5-: CTAGGCGAGGAGCTCCAGT -3 (Sigma-Aldrich). The PCR products were sequenced using the CEQ 8000 Genetic Analysis System (Beckman Coulter) and the analysis of the bidirectional sequence traces was performed using Seq Man Pro (DNASTAR, Inc) using as a reference the genomic sequence of the IDH2 gene (NM_002168).17,18
All patients were stratified in favorable, intermediate or adverse prognostic subgroup according to ELN risk stratification system 2017.4 The baseline characteristics of the patients, such as age and gender, FAB subtype, karyotype, risk stratification subgroup, NPM1 mutation and FLT3 status were evaluated. IDH1/IDH2 in this study group were correlated with demographic characteristics of the patients, FAB subtype, conventional cytogenetic findings, the risk stratification group, NPM1 and FLT3 status.
Furthermore, in patients that received intensive chemotherapy with or without allo-SCT (instead of palliative care) for AML, including standard treatment “3+7” as induction therapy and cytarabine-containing regimens as consolidation, IDH mutation status was correlated with the occurrence of death during induction therapy, as well as, with the resistance of the disease to the treatment.
The impact of isolated IDH mutations has been extensively studied especially in patients with normal cytogenetics. In order to investigate the impact of the IDH status on RFS and OS, survival analysis was restricted to patients that treatment approach is not considered “standardized”. Therefore, survival analysis was studied to the group of patients that received standard chemotherapy for AML while not dying during induction therapy, whose disease was not characterized as primary refractory and were not intended for allogeneic stem cell transplantation a priori. The last category includes those patients who were scheduled for allogeneic stem cell transplantation at diagnosis and proceeded at first complete remission.
Relapse Free Survival (RFS) was defined as the time between disease diagnosis and relapse or death from any cause or last follow up. Overall Survivall (OS) was defined as the time between disease diagnosis and death from any cause or last follow up.
The Chi square test was used for correlations between categorical variables. Cox models were used to identify prognostic values. Kaplan-Meier method was used to estimate the distribution of RFS and OS. All statistical analyses were performed using SPSS package for Windows.
RESULTS
One hundred and twenty-six (126) patients diagnosed with Acute Myeloid Leukemia (AML) in one Greek Hematology Department from 1995 to 2010 were retrospectively analyzed. There was an equal distribution between the two genders (males 63/126 and females 63/126). 52% of patients were <60 years old (65/126) and 48% were ≥60 years old (61/126). M2 AML subtype according to the FAB classification, was the most frequent subtype (40%) excluding M3 subtype since that category was not studied. Normal karyotype was found in 62/126 (53% of patients), whereas 54/126 (47%) had cytogenetic abnormalities. Patients were stratified into risk groups according to the 2017 ELN recommendations, 30% (37/126), 56% (71/126), 14% (18/126) of patients were classified in the favorable, intermediate and adverse risk group respectively. NPM1 mutations were detected in 39/126 (31% of the study group), while FLT3-ITD mutations were detected in 24/126 (19%). The patients’ characteristics are summarized in Table 1.
IDH mutations were observed in 17/126 (13.5%). IDH1 mutations were detected in 6/126 (4.8%), IDH2 in 9/126 (7.1%) and IDH1 and IDH2 mutations (dual mutations) in 2/126 (1.6%). The R132H mutation was the most prevalent type of IDH1 mutation, while the R140Q was the most prevalent type of IDH2 mutation. The correlation of the IDH status with the patients’ characteristics and the corresponding prognostic values is summarized in Table 2.
The presence of IDH mutations was not associated neither with gender (male vs female, p=0.118) nor with age (<60 vs ≥60, p= 0.508). Additionally, IDH mutations were not correlated with any FAB subtype (p= 0.526)and were not associated with karyotype (p= 0.168). IDH mutations were associated with the presence of NPM1 mutation (IDH mutations were present in 10/39 of NPM1 mutated patients, whereas in NPM1 wild type patients, IDH mutations were present in 7/87, P=0.017). There is no association with the presence of FLT3-ITD mutations (IDH mutations were present in 3/24 of FLT3-ITD mutated patients (p=1.0). Also, there is no significant correlation between IDH status and risk stratification group. Furthermore, IDH mutations did not seem to correlate: a) with the occurrence of death during induction therapy (p=1.0) in patients that received standard treatment for AML, b) with the resistance of the disease in the treatment (p=1.0) in patients that received standard treatment for AML and that did not die during induction therapy.
Additionally, survival analysis in the subgroup of patients mentioned above, revealed that IDH status is not related significantly with RFS (Figure 1) or OS (Figure 2) in this patients’ subgroup (p=0.753 and p=0.852 respectively).
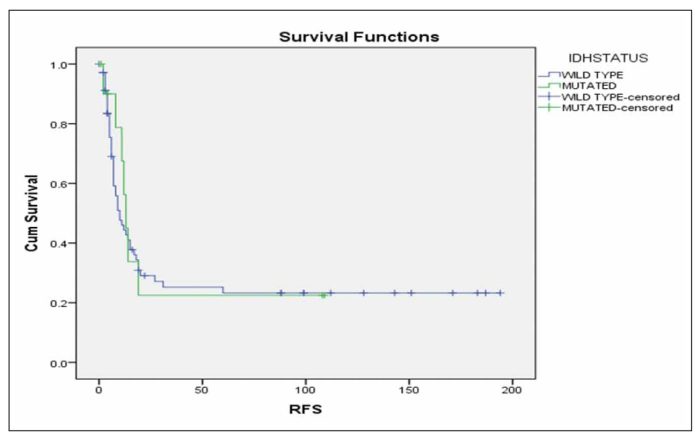
Figure 1

Figure 2

Figure 3
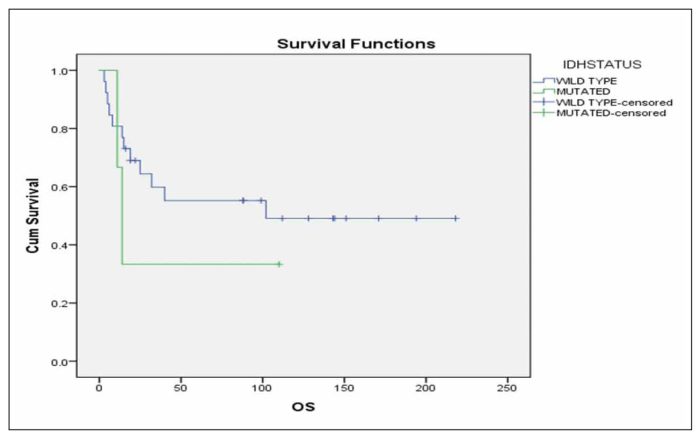
Figure 4

Figure 5

Figure 6

Figure 7
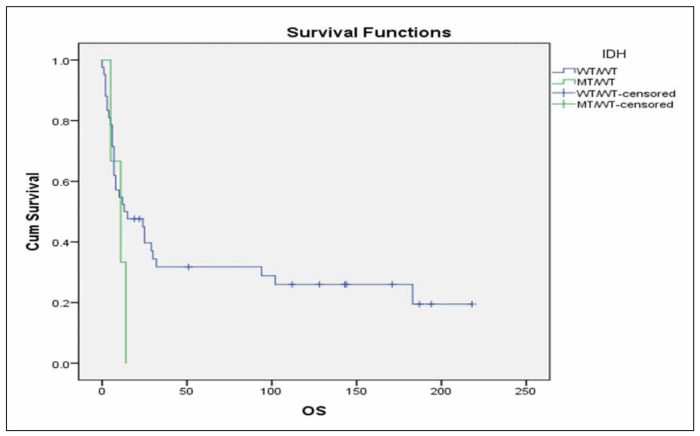
Figure 8
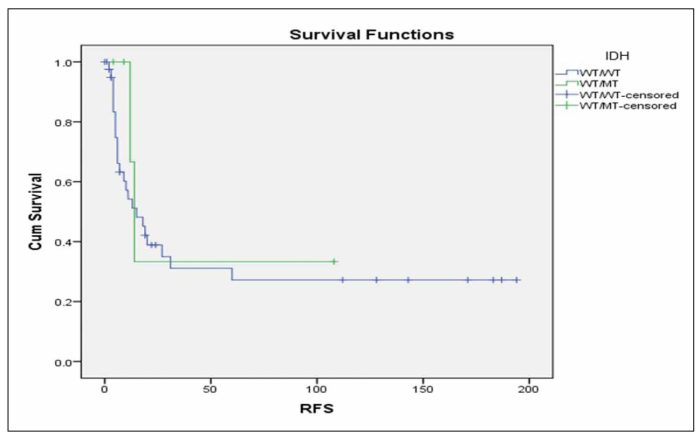
Figure 9
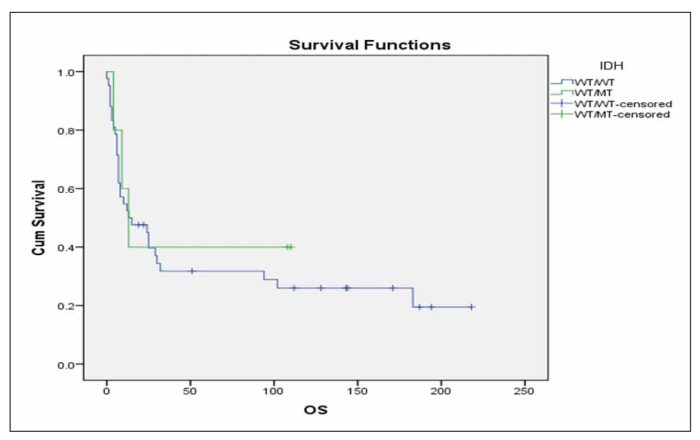
Figure 10

Figure 11
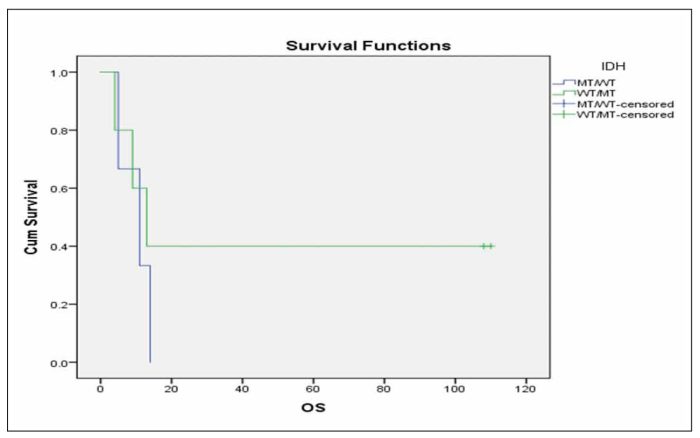
Figure 12
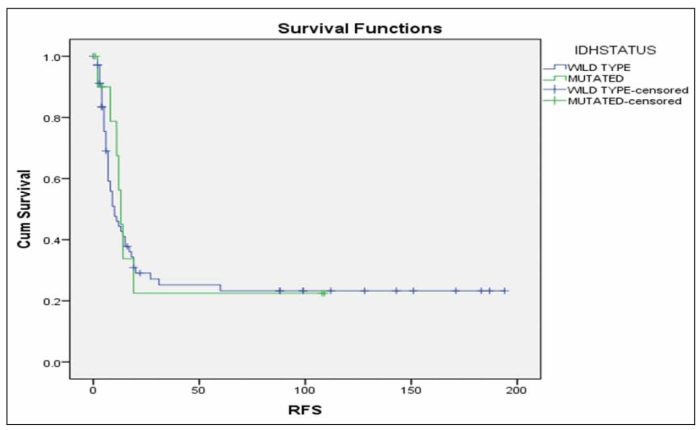
Figure 13
When we performed multivariate analysis FLT3-ITD and age ≥60 years were found to be independent prognostic factors for RFS and OS in this group of patients. Consequently, we investigated whether the IDH status was correlated with RFS or OS in the subgroup of patients with the aforementioned characteristics that were also FLT3 unmutated and <60 years old. It was found that patients with mutated IDH status have marginally inferior RFS in comparison with patients with unmutated IDH status (Figure 3, p=0.092), while there was no significant difference in terms of OS (Figure 4, p=0.491).We further analysedthe impact of isolated IDH mutations on RFS and OS and we observed that when IDH1 mutated patients compared with IDH2 mutated or unmutated IDH status patients, they seemed to have an inferior RFS (Figure 5, p=0.203) as well as OS (Figure 6, p=0.219), although not statistically significant. When we analysed only the subgroup of patients with IDH1 mutations, we observed that it seems have inferior outcome in terms of RFS (Figure 7, p=0.1) and OS (Figure 8, p=0.255), although the statistical significance is not significant. When we analysed only the subgroup of patients with IDH2 mutations we observed that it seems to have favorable outcome in terms of RFS (Figure 9, p=0.542) and OS (Figure 10, p=0.626), although the statistical significance failed to be demonstrated. When we compared IDH1 mutant patients to IDH2 ones, we observed significantly superior RFS (Figure 11, p=0.005) and but not statistically significant favorable outcome in OS (Figure 12, p=0.456) in IDH2 mutant patients.
We compared separately IDH2 mutated patients vs wild-type and we observed that patients with mutations IDH2 R172 and IDH2 R140Q presented the most favorable prognostic impact (Figure 13, p=0.188).
DISCUSSION
In this retrospective study of οne hundred and twenty-six 126 newly diagnosed patients with AML, we evaluated the frequency of IDH mutations, their association with other patients’ characteristics, as well as, karyotypic and molecular data and additionally their impact on RFS and OS.
There was an equal distribution between the two genders and as far as the age is concerned, 52% of patients were <60 years old and 48% were ≥60 years old. Normal karyotype was found in 53% of patients, whereas 47% had cytogenetic abnormalities. Patients were stratified into risk groups according to the 2017 ELN recommendations, into favorable, intermediate and adverse risk group. NPM1 mutations were detected in 31% of the study group, while FLT3-ITD mutations were detected in 19%. IDH mutations were observed in 13.5%. IDH1 mutations were detected in 4.8%, whereas IDH2 mutations in 7.1% and IDH1 and IDH2 mutations (dual mutations) in 1.6%. The R132H mutation was the most prevalent type of IDH1 mutation, while the R140Q was the most prevalent type of IDH2 mutation.
Earlier studies of Pascha et al19 had shown that IDH mutations were associated with older age, lower WBC, higher platelets, cytogenetically normal (CN) AML and NPM1 mutations, in particular with the genotype of mutated NPM1 without FLT3-ITD. In patients with CN-AML with the latter genotype, IDH mutations adversely impacted RFS and OS, whereas outcome was not affected in patients with CN-AML who lacked this genotype. In CN-AML, multivariable analyses revealed a significant interaction between IDH mutation and the genotype of mutated NPM1 without FLT3-ITD (the adverse impact of IDH mutation was restricted to this patient subset). All but one IDH1 mutation caused substitutions of residue R132, IDH2 mutations caused changes of R140 or R172.
Patel et al20 showed that patients without FLT3-ITD mutations and with both mutant NPM1 and IDH represent a favorable-risk subset defined by a specific mutational genotype, whereas patients who were negative for FLT3-ITD mutations and had mutant NPM1 without concurrent IDH mutations had a much less favorable outcome — particularly if those patients had concurrent mutations associated with an unfavorable-risk profile.
Papaemmanuil et al21 have reported that although the number of patients in their IDH2R172 study subgroup was small, the long-term outcomes were broadly similar to those in patients with NPM1-mutated AML. Furthermore, they stated that IDH2 should be considered along with TP53, SRSF2, ASXL1, DNMT3A, for incorporation into prognostic guidelines because they are common and exert a strong influence on clinical outcomes.
Green et al22 reported in 2011 for the first time the favorable outcome associated with an IDH2R140 mutation. When the results were stratified according to NPM1 genotype, relapse was reduced in IDH2R140 cases both with and without an NPM1 mutation.
In our patients IDH mutations were not associated neither with gender nor with age and were not associated with karyotype. Additionally, there was no significant correlation between IDH status and risk stratification group. Furthermore, IDH mutations were not associated with the FLT3 status of our patients, however, they were associated with the presence of NPM1 mutation. Also, IDH mutations did not seem to correlate with the occurrence of death during induction therapy in patients that received standard treatment for AML or with the resistance of the disease in the treatment in patients that received standard treatment for AML and that did not die during induction therapy.
In our study we observed that IDH mutations did not seem to significantly affect RFS and OS, concerning the group of patients that received standard treatment (instead of palliative care) for AML, that did not die during induction therapy, that were not intended for allogeneic stem cell transplantation a priori and that their disease was not characterized as primary refractory. When we performed multivariate analysis, FLT3-ITD and age ≥60 years were found to be independent prognostic factors for RFS and OS in this group of patients. We further analysed the impact of isolated IDH mutations on RFS and OS, in subgroup of patients with the aforementioned characteristics, with normal karyotype, irrespective of age, and we observed that when IDH1 mutated patients compared to patients with IDH2 mutations or unmutated IDH status ones, they seemed to have an inferior RFS as well as OS, although not statistically significant. As far as patients with IDH2 mutations is concerned, we observed that it seems to have favorable outcome in terms of RFS and OS (Figure 10, p=0.626), although the statistical significance is not significant. When we compared only IDH1 mutant patients to IDH2 ones, we observed significantly superior RFS in IDH2 mutant patients. Additionally, when we compared IDH2 mutant vs wild-type patients, the most favorable prognostic impact was observed in IDH2 R172 mutant patients, followed by IDH2 R14OQ mutant ones.
In conclusion, although the frequencies of our study are relative small, however we could make the statement that we observed a positive impact of IDH2 R172 and IDH2 R14OQ mutations in our study group, statement in agreement with all above mentioned studies.
Screening for IDH1 and IDH2 mutations will be soon applied to AML patients at presentation or relapse, due to availability of recently FDA approved IDH1 inhibitor ivosidenib, as the first treatment of adult patients with relapsed/refractory AML and an IDH1 mutation, as well as adult patients with de novo IDH1 AML unfit for chemotherapy.22 Enasidenib, has recently been FDA approved, as the first oral IDH2 inhibitor, in relapsed AML patients carrying this mutation.23.24
Conflict of Interest
None.
REFERENCES
- Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015 Sep;73(12):1136-52.
- Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001 Sep;14(3):497-529.
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009 Jul;114(5):937-51.
- Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017 Jan;129(4):424-47.
- Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, et al. Medical research council adult leukaemia working party. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008 Mar;111(5):2776-84.
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002 Sep;100(5):1532-42.
- Frohling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: Pathogenetic and clinical implications. J Clin Oncol. 2005, 23:6285-6295.
- Dohner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European Leukemia-Net. Blood. 2010 Jan;115(3):453-74.
- Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179-98.
- Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. AMLCG Study Group Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016 Aug;128(5):686-98.
- Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017 Feb;31(2):272-81.
- Dinardo CD, Ravandi F, Agresta S, Konopleva M, Takahashi K, Kadia T, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015 Aug;90(8):732-6.
- Fernandez-Mercado M, Yip BH, PellagattiA, Davies C, José Larrayoz M, Kondo T, et al. Mutation patterns of 16 genes in primary and secondary acute myeloid leukemia (AML) with normal cytogenetics. PLoS ONE [Internet]. 2012 Aug;e42334 2012. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0042334.
- Winer J, Jung CK, Shackel I, et al. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999 May;270(1):41-9.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977 Dec;74(12):5463-7.
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, et al. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. Nature. 1977 Feb 24;265(5596):687-95.
- ES Lander, LM Linton, B Birren, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb;409(6822):860-921.
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010 Aug;28(22):3636-43.
- Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Levine, prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012 Mar;366(12):1079-89.
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl J Med. 2016 Jun;374(23):2209-21.
- Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC, et al. The prognostic significance of IDH2 mutations inAMLdepends on the location of the mutation. Blood. 2011 Jul;118(2):409-12.
- Golub D, Iyengar N, Dogra S, Wong T, Bready D, Tang K, et al. Placantonakis mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front Oncol. 2019 May;9:417.
- Amaya ML, Pollyea DA. Targeting the IDH2 pathway in acute myeloid leukemia. Clin Cancer Res. 2018 Oct;24(20):4931-36.